New Paper: Human Gut Microbiota Predicts Susceptibility to Vibrio cholerae infection
Press Release
Duke Today. AI Detects Patterns of Gut Microbes for Cholera Risk. [May 7, 2018] by Marla Vacek Broadfoot.
Paper Details
FS Midani\(\dagger\), AA Weil\(\dagger\), F Chowdhury, YA Begu, AI Khan, M Debela, HK Durand, AT Reese, SN Nimmagadda, JD Silverman, CN Ellis, ET Ryan, SB Calderwood, JB Harris, F Qadri, LA David\(\ddagger\), and RC LaRocque\(\ddagger\).
Human Gut Microbiota Predicts Susceptibility to Vibrio cholerae Infection
The Journal of Infectious Diseases. 2018. https://doi.org/10.1093/infdis/jiy192
Background
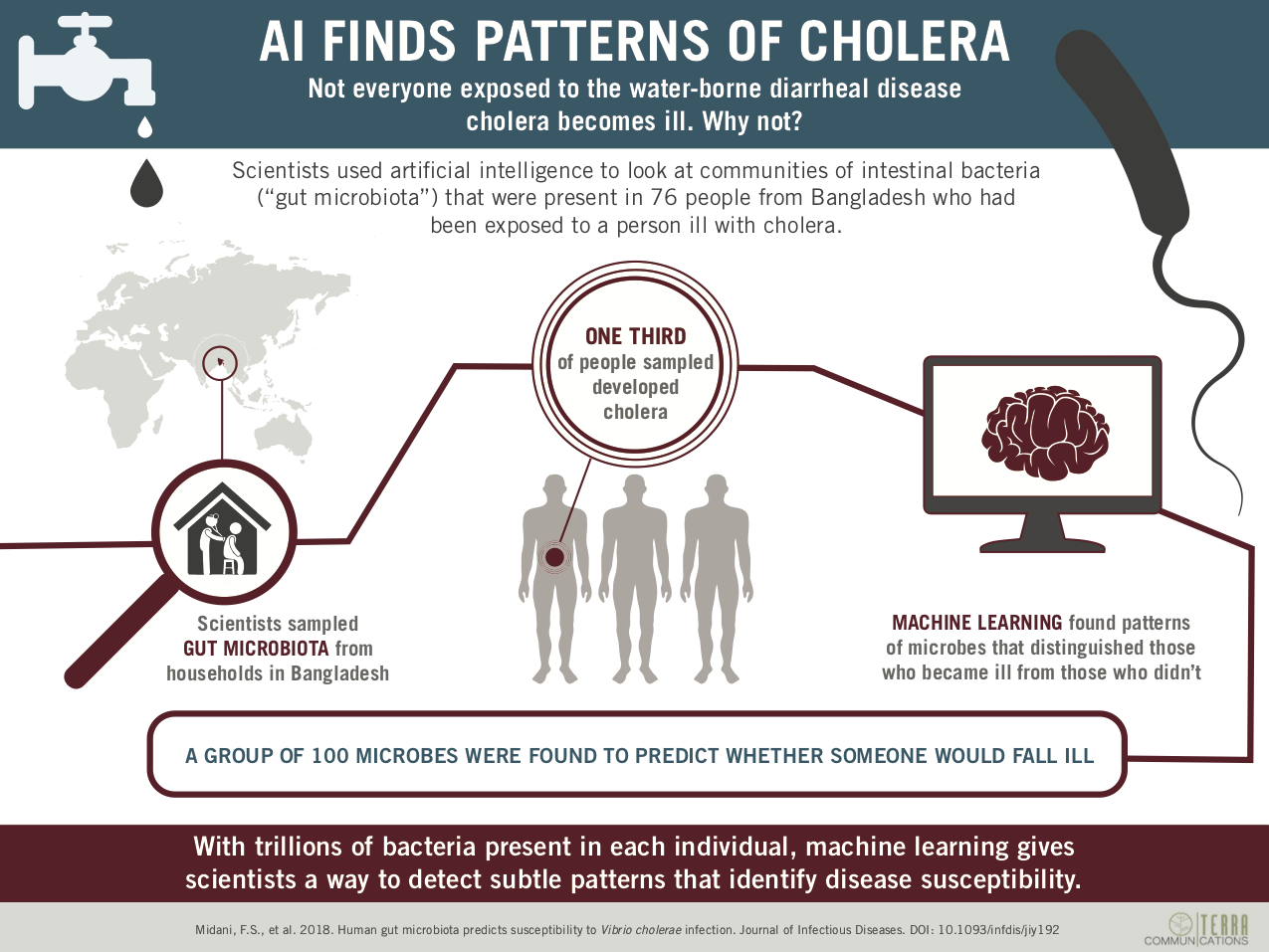
Cholera is a public health problem worldwide and the risk factors for infection are only partially understood.
Methods
We prospectively studied household contacts of cholera patients to compare those who were infected with those who were not. We constructed predictive machine learning models of susceptibility using baseline gut microbiota data. We identified bacterial taxa associated with susceptibility to Vibrio cholerae infection and tested these taxa for interactions with V. cholerae in vitro.
Results
We found that machine learning models based on gut microbiota predicted V. cholerae infection as well as models based on known clinical and epidemiological risk factors. A ‘predictive gut microbiota’ of roughly 100 bacterial taxa discriminated between contacts who developed infection and those who did not. Susceptibility to cholera was associated with depleted levels of microbes from the phylum Bacteroidetes. By contrast, a microbe associated with cholera by our modeling framework, Paracoccus aminovorans, promoted the in vitro growth of V. cholerae. Gut microbiota structure, clinical outcome, and age were also linked.
Conclusion
These findings support the hypothesis that abnormal gut microbial communities are a host factor related to V. cholerae susceptibility.
Data & Code Sharing
Raw Data of nucleotide sequences used in this study can be downloaded from the European Nucleotide Archive under accession number PRJEB17860.
Code for deriving data, running classifiers, plotting figures, and tabulating data are provided here on GitHub.
