New Software: AMiGA for the Analysis of Microbial Growth Assays
Check out my new software AMiGA (see code and documentation) for analyzing microbial growth curves. At the @BrittonTML, we had a pretty good system for analyzing microbial growth especially on Biolog PM plates, but we focused on a single metric which missed some intereting growth dynamics.
I started with published tools and writing my own code to fit growth curves using classical models like Gompertz but quickly realized that clinical isoaltes including Clostridioides difficile are quite finicky and have varying shapes and growth dynamics.
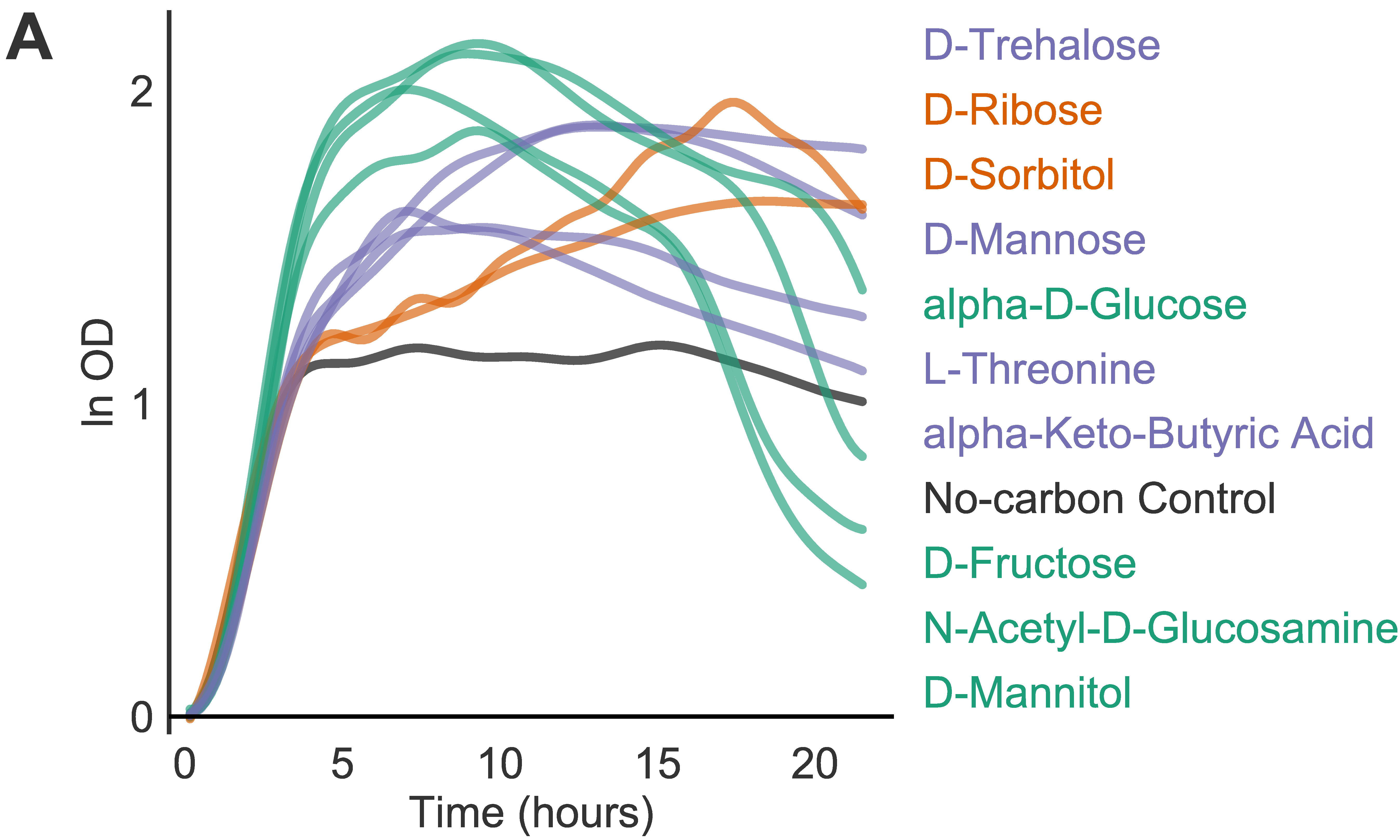
To get around this issue, I adopted a fairly nascent approach of modelling growth curves with Gaussian Proces regression. It worked really well, and the rest of the team and collaborators became interested in this approach.
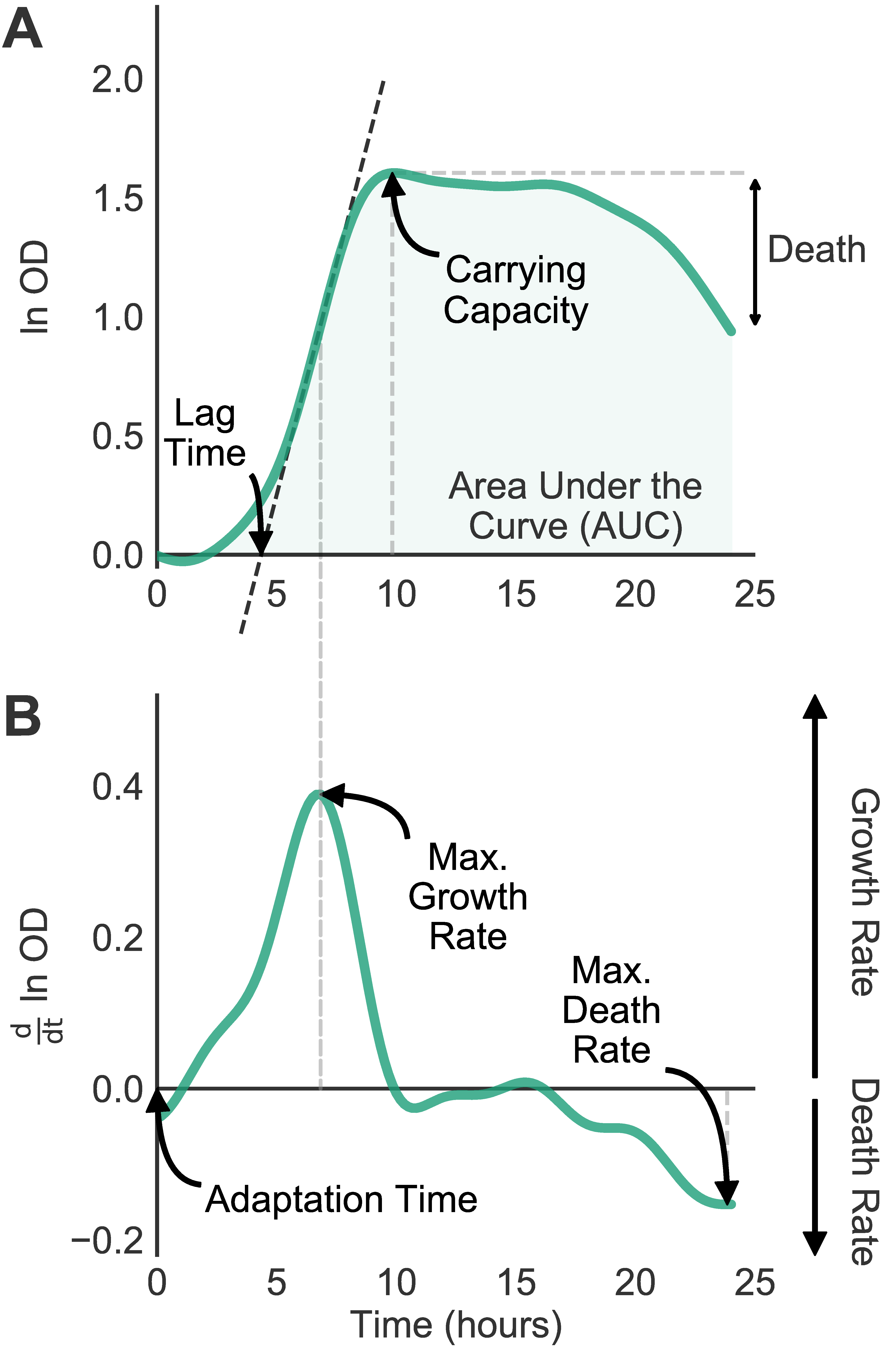
So I took my scripts and turned them into a user-friendly program Analysis of Microbial Growth Asssays (AMiGA). I spent lots of time on documentation and examples, see https://firasmidani.github.io/amiga.

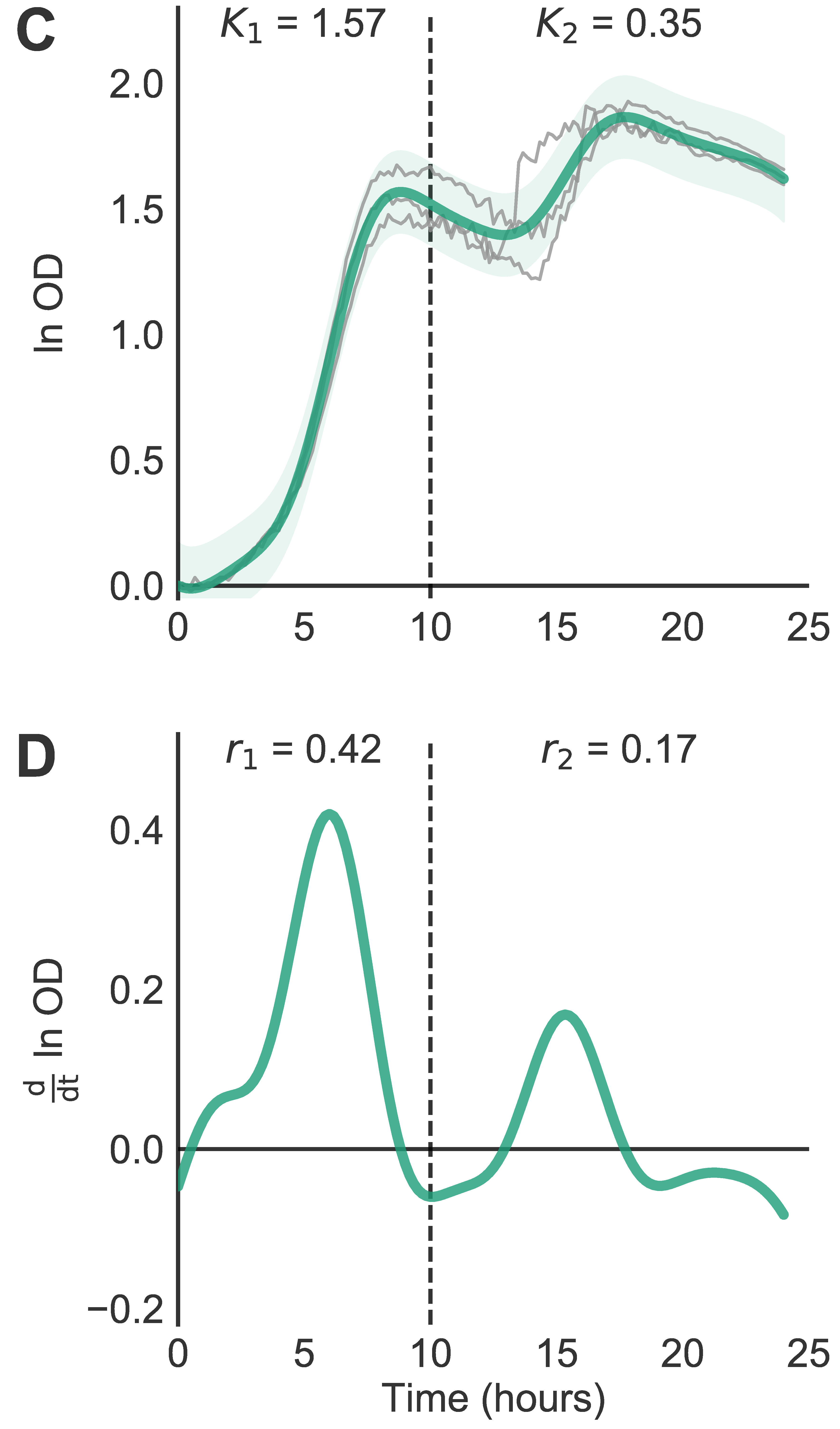
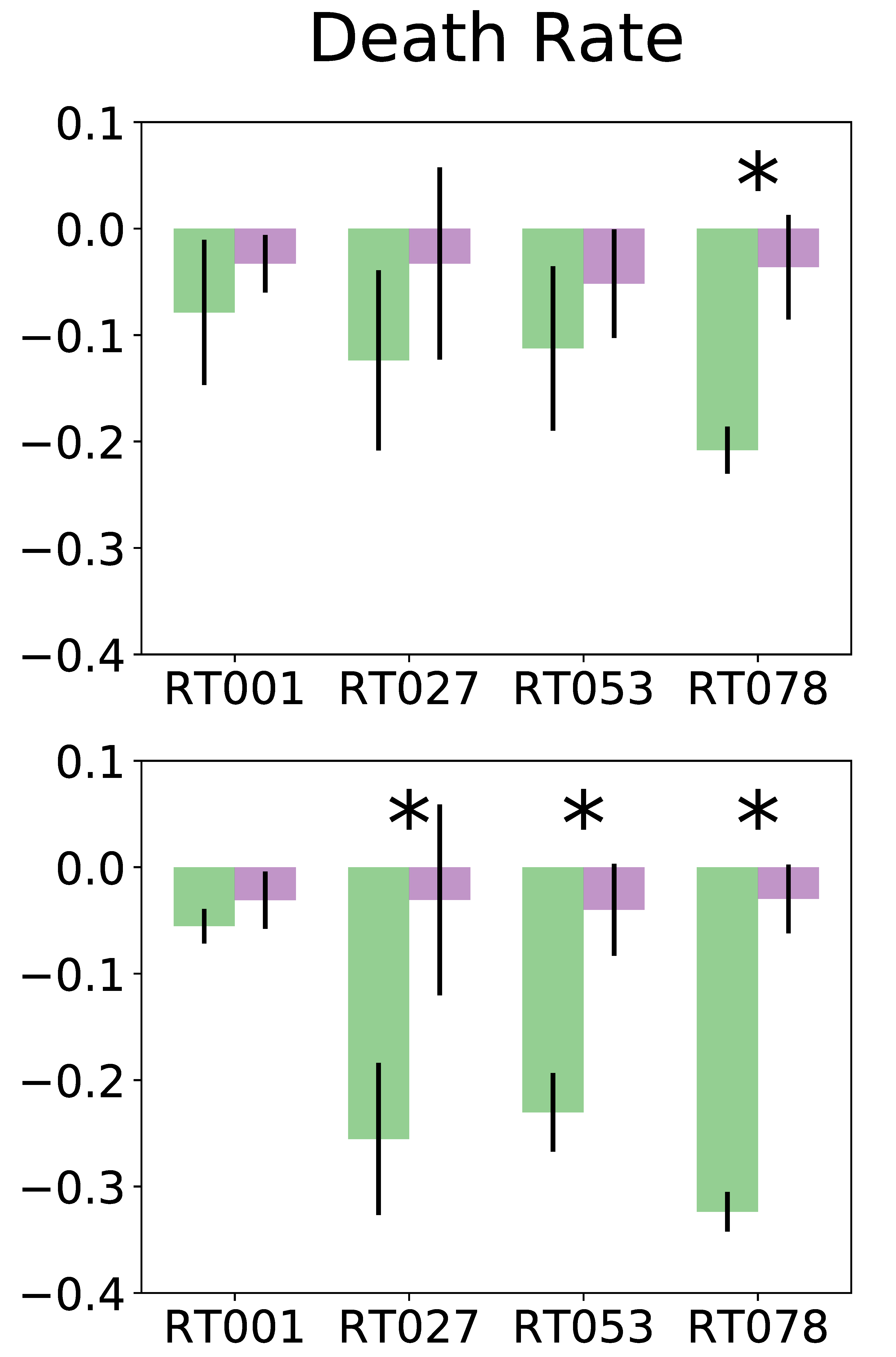
AMiGA can fit individual curves or pool replicates (from different plates) and give you confidence intervals too. It can estimate death rates; and detect & characterize diauxic shifts. I’m not aware of any existing software that automatically quantifies eitehr phenotypes, so feedback welcome.
Users can interact with AMiGA via the command-lien (terminal). It requires minimal and often no pre-processing of your raw data.
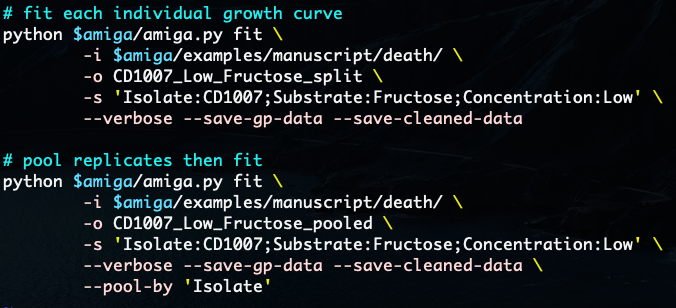
You can also throw in some meta-data to test hypotheses of differential growth. Feel free to report bugs or ask questions on email or GitHub repo.
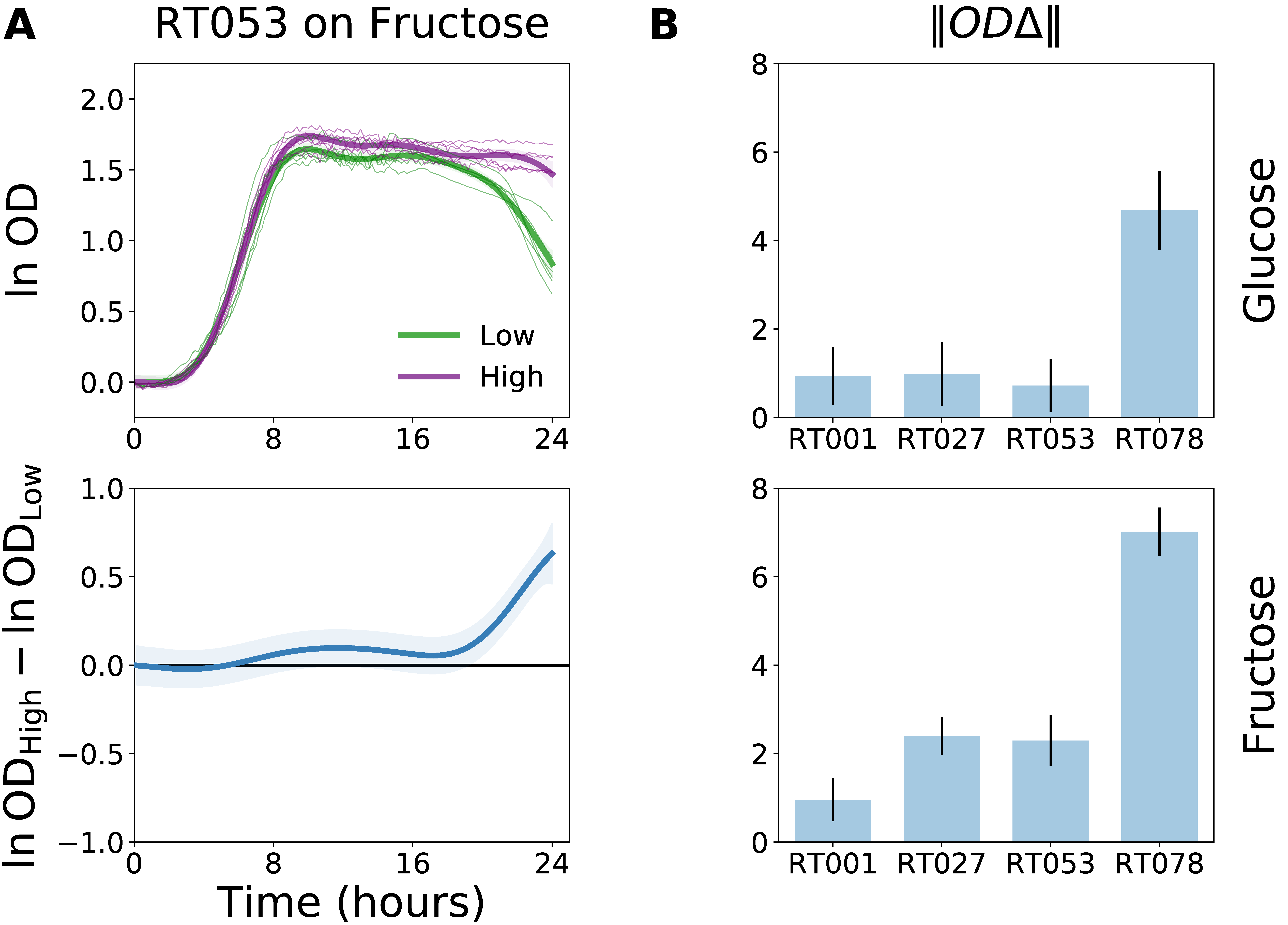
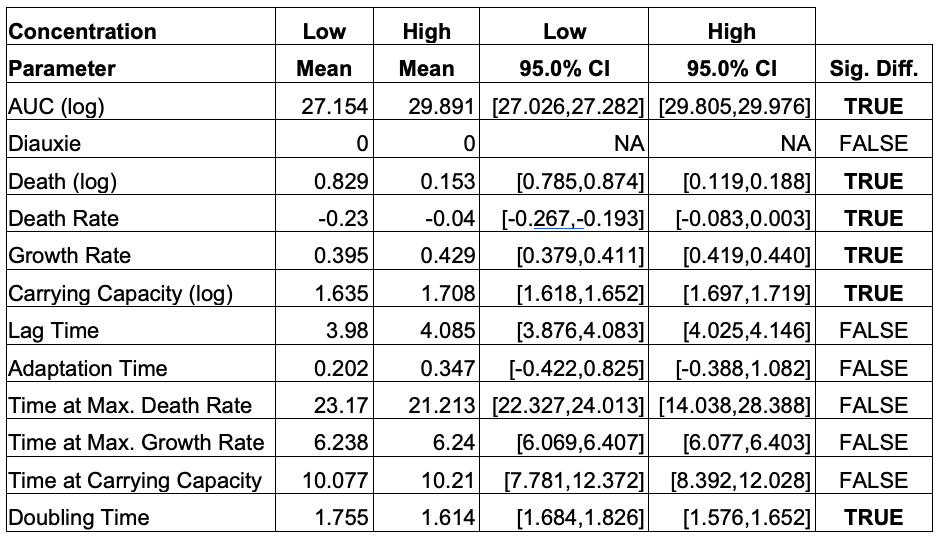
Paper Details
FS Midani, J Collins, and RA Britton.
AMiGA: software for the Analysis of Microbial Growth Assays
bioRxiv. 2020. https://www.biorxiv.org/content/10.1101/2020.11.04.369140v1
Abstract
The analysis of microbial growth is one of the central methods in the field of microbiology. Microbial growth dynamics can be characterized by growth parameters including carrying capacity, exponential growth rate, and growth lag. However, growth assays with clinical isolates, fastidious organisms, or microbes under stress often produce atypical growth shapes that do not follow the classical microbial growth pattern. Here, we introduce the Analysis of Microbial Growth Assays (AMiGA) software which streamlines the analysis of growth curves without any assumptions about their shapes. AMiGA can pool replicates of growth curves and infer summary statistics for biologically meaningful growth parameters. In addition, AMiGA can quantify death phases and characterize diauxic shifts. It can also statistically test for differential growth under distinct experimental conditions. Altogether, AMiGA streamlines the organization, analysis, and visualization of microbial growth assays.
Importance
Our current understanding of microbial physiology relies on the simple method of measuring microbial populations’ size over time and under different conditions. Many advances have increased the throughput of those assays and enabled the study of non-lab adapted microbes under diverse conditions that widely affect their growth dynamics. Our software provides an all-in-one tool for estimating the growth parameters of microbial cultures and testing for differential growth in a high-throughput and user-friendly fashion without any underlying assumptions about how microbes respond to their growth conditions.
